Bacteria grow in communities of many co-occurring species in , e.g., in your gut, in soil, or in the ocean. A fundamental process in these communities (more specifically, communities of heterotrophic bacteria, i.e., bacteria that utilize organic carbon sources) is that bacteria take up substrates (basically, food) like sugars and amino acids from the environment and turn them into biomass or convert them into something else they then excrete. For this new paper, what we wanted to know was: which substrates can different bacteria use (we were focused on substrates they can use as a carbon source)? Can we identify patterns of substrate utilization, e.g., are similar compounds consumed by similar bacteria? Can we predict these patterns by looking at which genes different bacteria encode? Our work touches on several important questions in microbiology, from microbial ecology (how do microbial communities work?) to biochemistry (how does the structure of metabolic pathways shape substrate utilization patterns?) to genomics & evolution (how are capabilities of substrate utilization encoded in the genome, and how did evolution shape these genomic patterns?).
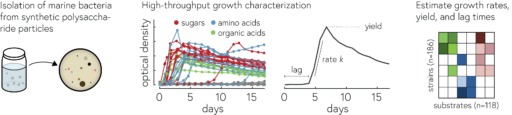
By analyzing the growth of 182 different strains of marine bacteria on 135 different potential carbon sources, we found that we can describe the substrate preferences of our bacteria to a first approximation in terms of their preference for sugars (e.g., glucose or polysaccharides like starch) relative to acids (e.g., amino acids or organic acids which are important intermediate during the chemical conversion of substrates into biomass). This preference is encoded in the genomes of bacteria, which tells us about the evolution of these preferences, but also makes the preferences predictable from genomes.
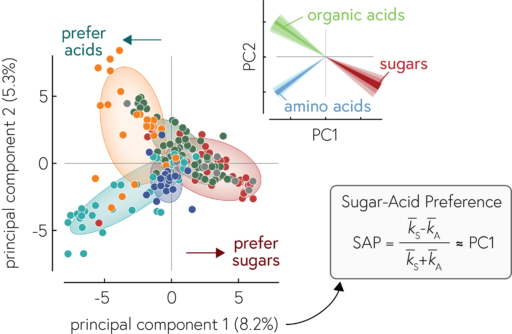
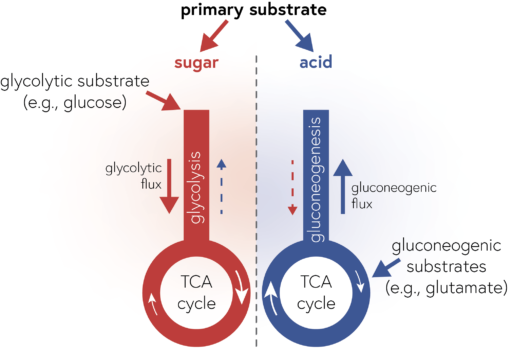
Our work reveals a way to simplify how we think about the metabolic capabilities of bacteria: we can describe a given heterotrophic bacterium by its degree of specialization along the axis of sugar to acid specialists. This is very useful because it allows us to describe communities of bacteria in a simple way (e.g., by their collective degree of specialization). More fundamentally, our work also shows how the evolution of bacterial genomes is structured by biochemical constraints which drives bacteria to specialize along this axis of sugar to acid specialists. Since the metabolic preferences are encoded in genomes, we can estimate the metabolic capabilities of species that we have not (yet) cultured, but for which we have genomic information (e.g., by sequencing entire communities and piecing together the genomes of the constituent species, a process called metagenomics). This allows us to begin to understand the metabolic processes in many microbial communities in the environment in a simplified manner.
Out now: Genome content predicts the carbon catabolic preferences of heterotrophic bacteria


Recent Comments