Evelina Tutucci

Research Topics
A fascinating aspect of all cells is their complex intracellular organization. Cells are subdivided in different compartments to organize cellular functions by physically separating biological reactions.
This compartmentalization is fundamental to regulate gene expression both in space and time. To understand how cells dynamically control gene expression to adapt to changing environments, we use single molecule mRNA imaging technologies both for fixed and living cells. These methods allow us to follow the entire mRNA life, from transcription to degradation, from the nucleus to the cytoplasm. Using these tools, we measure, with high spatial and temporal resolution, each stage of gene expression in individual cells. We study the coordination between the different stages of gene expression, to understand how cells tune gene expression to improve their fitness. By looking at single cells, we can assess how gene expression noise and cell-to cell variability affect cellular functions.
Using cell cycle mRNA as a paradigm and S. cerevisiae as a model system, we study how cells coordinate mRNA localization, translation and decay to achieve precise gene expression regulation.
For more information visit my personal website www.tutuccilab.com
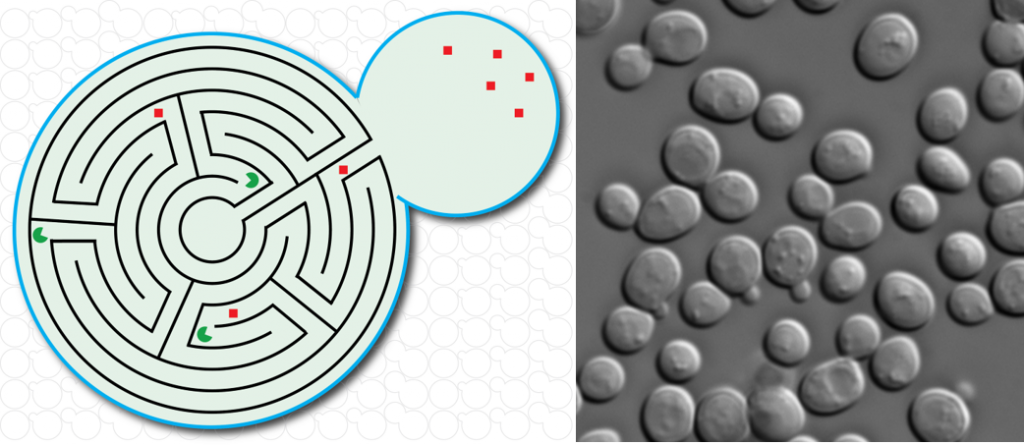
Project 1 – Methods development for mRNA imaging in fixed and living cells
To study gene expression in single cells, we continuously develop fluorescence-based imaging tools to visualize single mRNAs in fixed and living cells.
For fixed cells, we developed a single molecule fluorescent in situ hybridization protocol combined to immuno-fluorescence for S. cerevisiae.
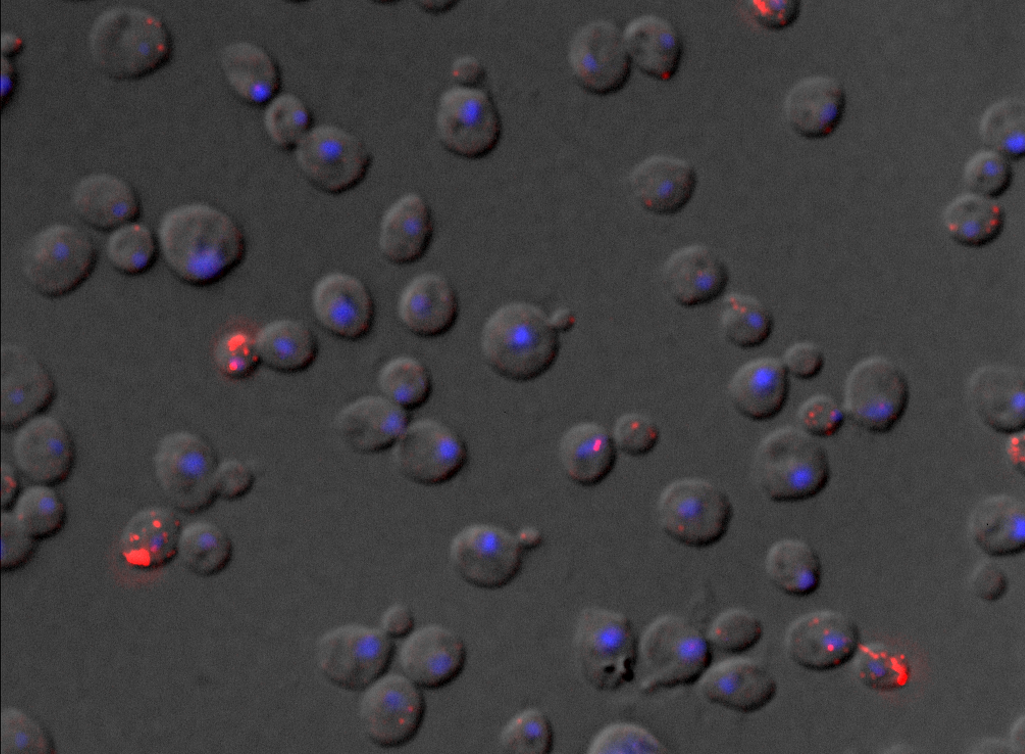
For living cells, we previously developed a new RNA tagging system (MS2V6) that allows us to tag individual mRNAs in S. cerevisiae and that, compared to previous reporters is particularly suited to tag unstable mRNAs. See: https://doi.org/10.1038/nmeth.4502 and https://doi.org/10.1038/s41596-018-0037-2
Project 2 – Spatial and Temporal regulation of gene expression in single cells
We study how the mRNA localization, translation and decay are coordinated within individual cell, by combining imaging, molecular biology, genetics and biochemistry.
We are particularly interested in understanding how cell cycle regulated genes such as the ASH1 mRNA tune their expression to control growth and maximize fitness.
Contact information
Email: evelina.tutucci@vu.nl
Personal website: www.tutuccilab.com
Systems Biology Lab
Vrije Universiteit Amsterdam
Room O|2 01E57
De Boelelaan 1108
1081 HZ Amsterdam
EDUCATION
- 2008-2013 Doctor of Philosophy, Molecular and Cellular Biology, University of Geneva, Switzerland.
- 2008 Master of Science, Medical and Pharmaceutical Biotechnology, University of Genoa, Italy.
- 2007 Bachelor of Science, Biotechnology, University of Genoa, Italy.
PROFESSIONAL EXPERIENCE
- December 2022 – Present : Assistant Professor, Systems Biology Lab, Vrije University Amsterdam, Netherlands.
- August 2019 – November 2022: Tenure-track Assistant Professor, Systems Biology Lab, Vrije University Amsterdam, Netherlands.
- 2014 – 2019: Postdoctoral fellow, Laboratory of Dr. Robert H. Singer Department of Anatomy and Structural Biology, Albert Einstein College of Medicine, Bronx, NY.
- 2008 – 2014: Graduate Student, Laboratory of Dr. Françoise Stutz, Department of Cellular of Geneva, Switzerland.
- 2005 – 2008: Undergraduate work, Laboratory of Dr. Paolo Malatesta Department Of Developmental Biology, University of Genoa, Italy.
FELLOWSHIPS AND AWARDS
- 2018 Travel award the RNA Society.
- 2017 Travel fellowship Swiss National Science Foundation: RNA society meeting, Prague.
- 2017 Travel award by the RNA Society .
- 2017 Poster prize RNA meeting 2017, RNA society.
- 2016 Advanced Postdoctoral Fellowship, Swiss National Science Foundation, Dept. Anatomy and Structural Biology, Albert Einstein College of Medicine, USA
- 2015 Travel fellowship Swiss National Science Foundation: Eukaryotic mRNA processing meeting.
- 2019 Travel award by the RNA Society .
- 2014 Early Postdoctoral Fellowship, Swiss National Science Foundation, Dept. Anatomy and Structural Biology, Albert Einstein College of Medicine, USA
- 2011 Travel award by the RNA Society .
- 2011 Travel award to attend Mechanisms of Nucleocytoplasmic Trafficking, EMBO.
PATENTS
Robert H. Singer, Evelina Tutucci and Maria Vera “RNA TAGGING SYSTEM FOR VISUALIZATION OF SINGLE mRNA MOLECULES” Provisional application to the US Patent and Trademark Office (no. 62/487,058).
PEER-REVIEWED PUBLICATIONS
- Botman, D, Kanagasabapathi, S, Rep, MI, van Rossum, K, Tutucci, E, Teusink, B et al.. cAMP in budding yeast: Also a messenger for sucrose metabolism?. Biochim Biophys Acta Mol Cell Res. 2024;1871 (4):119706. doi: 10.1016/j.bbamcr.2024.119706. PubMed PMID:38521467 .
- van Otterdijk, S, Motealleh, M, Wang, Z, Visser, TD, Savakis, P, Tutucci, E et al.. Single-Molecule Fluorescent In Situ Hybridization (smFISH) for RNA Detection in the Fungal Pathogen Candida albicans. Methods Mol Biol. 2024;2784 :25-44. doi: 10.1007/978-1-0716-3766-1_2. PubMed PMID:38502476 .
- Gerber, A, van Otterdijk, S, Bruggeman, FJ, Tutucci, E. Understanding spatiotemporal coupling of gene expression using single molecule RNA imaging technologies. Transcription. 2023;14 (3-5):105-126. doi: 10.1080/21541264.2023.2199669. PubMed PMID:37050882 PubMed Central PMC10807504.
- Das, S, Vera, M, Gandin, V, Singer, RH, Tutucci, E. Author Correction: Intracellular mRNA transport and localized translation. Nat Rev Mol Cell Biol. 2021;22 (7):505. doi: 10.1038/s41580-021-00374-6. PubMed PMID:33911234 .
- Das, S, Vera, M, Gandin, V, Singer, RH, Tutucci, E. Intracellular mRNA transport and localized translation. Nat Rev Mol Cell Biol. 2021;22 (7):483-504. doi: 10.1038/s41580-021-00356-8. PubMed PMID:33837370 PubMed Central PMC9346928.
- Tsuboi, T, Viana, MP, Xu, F, Yu, J, Chanchani, R, Arceo, XG et al.. Mitochondrial volume fraction and translation duration impact mitochondrial mRNA localization and protein synthesis. Elife. 2020;9 :. doi: 10.7554/eLife.57814. PubMed PMID:32762840 PubMed Central PMC7413667.
- Pichon, X, Robert, MC, Bertrand, E, Singer, RH, Tutucci, E. New Generations of MS2 Variants and MCP Fusions to Detect Single mRNAs in Living Eukaryotic Cells. Methods Mol Biol. 2020;2166 :121-144. doi: 10.1007/978-1-0716-0712-1_7. PubMed PMID:32710406 PubMed Central PMC8950302.
- Tutucci, E, Singer, RH. Simultaneous Detection of mRNA and Protein in S. cerevisiae by Single-Molecule FISH and Immunofluorescence. Methods Mol Biol. 2020;2166 :51-69. doi: 10.1007/978-1-0716-0712-1_4. PubMed PMID:32710403 .
- Maekiniemi, A, Singer, RH, Tutucci, E. Single molecule mRNA fluorescent in situ hybridization combined with immunofluorescence in S. cerevisiae: Dataset and quantification. Data Brief. 2020;30 :105511. doi: 10.1016/j.dib.2020.105511. PubMed PMID:32368581 PubMed Central PMC7186551.
- Vera, M, Tutucci, E, Singer, RH. Imaging Single mRNA Molecules in Mammalian Cells Using an Optimized MS2-MCP System. Methods Mol Biol. 2019;2038 :3-20. doi: 10.1007/978-1-4939-9674-2_1. PubMed PMID:31407274 PubMed Central PMC8950109.
- Infantino, V, Tutucci, E, Yeh Martin, N, Zihlmann, A, Garcia-Molinero, V, Silvano, G et al.. The mRNA export adaptor Yra1 contributes to DNA double-strand break repair through its C-box domain. PLoS One. 2019;14 (4):e0206336. doi: 10.1371/journal.pone.0206336. PubMed PMID:30951522 PubMed Central PMC6450643.
- Tutucci, E, Vera, M, Singer, RH. Single-mRNA detection in living S. cerevisiae using a re-engineered MS2 system. Nat Protoc. 2018;13 (10):2268-2296. doi: 10.1038/s41596-018-0037-2. PubMed PMID:30218101 .
- Tutucci, E, Livingston, NM, Singer, RH, Wu, B. Imaging mRNA In Vivo, from Birth to Death. Annu Rev Biophys. 2018;47 :85-106. doi: 10.1146/annurev-biophys-070317-033037. PubMed PMID:29345990 .
- Tutucci, E, Vera, M, Biswas, J, Garcia, J, Parker, R, Singer, RH et al.. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat Methods. 2018;15 (1):81-89. doi: 10.1038/nmeth.4502. PubMed PMID:29131164 PubMed Central PMC5843578.
- Brickner, DG, Sood, V, Tutucci, E, Coukos, R, Viets, K, Singer, RH et al.. Subnuclear positioning and interchromosomal clustering of the GAL1-10 locus are controlled by separable, interdependent mechanisms. Mol Biol Cell. 2016;27 (19):2980-93. doi: 10.1091/mbc.E16-03-0174. PubMed PMID:27489341 PubMed Central PMC5042583.
- Wu, B, Miskolci, V, Sato, H, Tutucci, E, Kenworthy, CA, Donnelly, SK et al.. Synonymous modification results in high-fidelity gene expression of repetitive protein and nucleotide sequences. Genes Dev. 2015;29 (8):876-86. doi: 10.1101/gad.259358.115. PubMed PMID:25877922 PubMed Central PMC4403262.
- Infante, T, Cesario, E, Gallo, M, Fazioli, F, De Chiara, A, Tutucci, C et al.. Ex vivo behaviour of human bone tumor endothelial cells. Cancers (Basel). 2013;5 (2):404-17. doi: 10.3390/cancers5020404. PubMed PMID:24216983 PubMed Central PMC3730325.
- Tutucci, E, Stutz, F. Keeping mRNPs in check during assembly and nuclear export. Nat Rev Mol Cell Biol. 2011;12 (6):377-84. doi: 10.1038/nrm3119. PubMed PMID:21602906 .
- Terrile, M, Appolloni, I, Calzolari, F, Perris, R, Tutucci, E, Malatesta, P et al.. PDGF-B-driven gliomagenesis can occur in the absence of the proteoglycan NG2. BMC Cancer. 2010;10 :550. doi: 10.1186/1471-2407-10-550. PubMed PMID:20939912 PubMed Central PMC2964636.
- Iglesias, N, Tutucci, E, Gwizdek, C, Vinciguerra, P, Von Dach, E, Corbett, AH et al.. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 2010;24 (17):1927-38. doi: 10.1101/gad.583310. PubMed PMID:20810649 PubMed Central PMC2932974.
PRE-PRINTS
1. Infantino V*, Tutucci E*, Bagdiul I, Palancade B, Yeh Martin N, Zihlman A, Garcia-Molinero V, Silvano G and Stutz F. The mRNA export adaptor Yra1 contributes to DNA double-strand break repair through its C-box domain (*Equal contribution). BioRxiv (October 15th, 2018). https://doi.org/10.1101/441980
2. Tsuboi T, Viana MP, Xu F, Yu J, Chanchani R, Arceo XG, Tutucci E, Choi J, Chen YS, Singer RH, Rafelski SM, Zid BM. Mitochondrial volume fraction controls translation of nuclear-encoded mitochondrial proteins. BioRxiv (January 1st, 2019) https://doi.org/10.1101/529289
3. Tutucci E*, Maekiniemi A, Snoep SL, Seiler M, van Rossum K, van Niekerk DD, Savakis P, Zarnack K, Singer RH*. Cyclin CLB2 mRNA localization determines efficient protein synthesis to orchestrate bud growth and cell cycle progression. BioRxiv (March 2nd, 2022)doi: 10.1101/2022.03.01.481833 (*Corresponding authors)
PUBLISHED DATASETS
1. Tutucci E, Maekiniemi A and Singer RH. (2020), “Single molecule mRNA Fluorescent In Situ Hybridization combined to Immunofluorescence in S. cerevisiae: Dataset and quantification”, Mendeley Data, v4 http://dx.doi.org/10.17632/bcmn9cxyzs.4
For an updated list of publications see my Google Scholar web page: https://scholar.google.com/citations?user=AbW4QQ0AAAAJ&hl=en
To download the pdf files go to https://www.tutuccilab.com/publications
Supervision of students (BSc and MSc)
For an updated list go to https://www.tutuccilab.com/team

Recent Comments