A genome-scale metabolic reconstruction is a network of all biochemical reactions that can occur in a cell. These reconstructions have proven to be very useful because they can be used to make phenotypic predictions. Many computational tools to create these reconstructions have been created (or updated) recently, making these genome-scale reconstructions available for everyone. However,
Lab News
In press: Dennis’ extensive characterisation of Fluorescent protein behaviour in yeast is published!
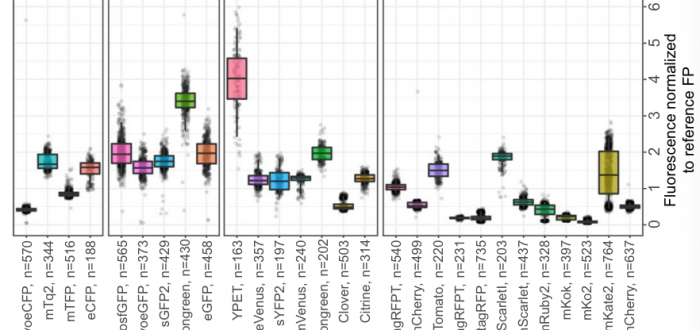
Dennis’ work on the behaviour of fluorescent proteins (FPs) in baker’s yeast has been published! With the help of Daan de Groot and Phillipp Schmidt, a variety of fluorescent proteins were characterised for various properties, including brightness, photobleaching, photochromism, day-to-day variation and monomerism. Many FPs showed different performance in yeast compared to previous characterisations done





Recent Comments